Western blot
Western blot is a protein detection analysis. Proteins are isolated from a sample and thereafter separated by gel electrophorsis. The proteins are then transferred from the gel onto a membrane with affinity for proteins, often nitrocellulose or polyvinulidene difluoride (PVDF) membranes. The transfer is called electroblotting, as the gel and membrane are placed into a chamber filled with a specific buffer as an electric current travels through the chamber. this moves the negatively charged proteins towards the positive anode and the proteins end-up on the membrane. Proteins can then be visualised by incubating the membrane with first a primary antibody (an antibody that attaches to the protein of interest) and then a fluorescently tagged secondary antibody is added to the mix, attaching only to the primary antibody. The protein of interest can then be identified by exciting the fluorescently tagged antibody which in turn emits a light for a camera to detect. To be able to quantify the amount of protein that is present on the membrane a secondary protein of is labeled as well in the incubation as a control. This is generaly a protein that has stable expression within the sample. This way we can determine the relative amount of the protein of interest to the control protein by dividing the light intensities of both of the proteins.
Pros: Can tell us if a protein is present or not in
the sample.
Cons: Only gives us the relative amount of protein present to another protein, and therefore can only answer if there is more protein present or less compared to another sample. And if the amount is similar it might lead to subjective results.
Examples of uses in the field
~ to detect anti-HIV antibody in human serum sample.
~ to detect prion proteins in a sample.
Cons: Only gives us the relative amount of protein present to another protein, and therefore can only answer if there is more protein present or less compared to another sample. And if the amount is similar it might lead to subjective results.
Examples of uses in the field
~ to detect anti-HIV antibody in human serum sample.
~ to detect prion proteins in a sample.
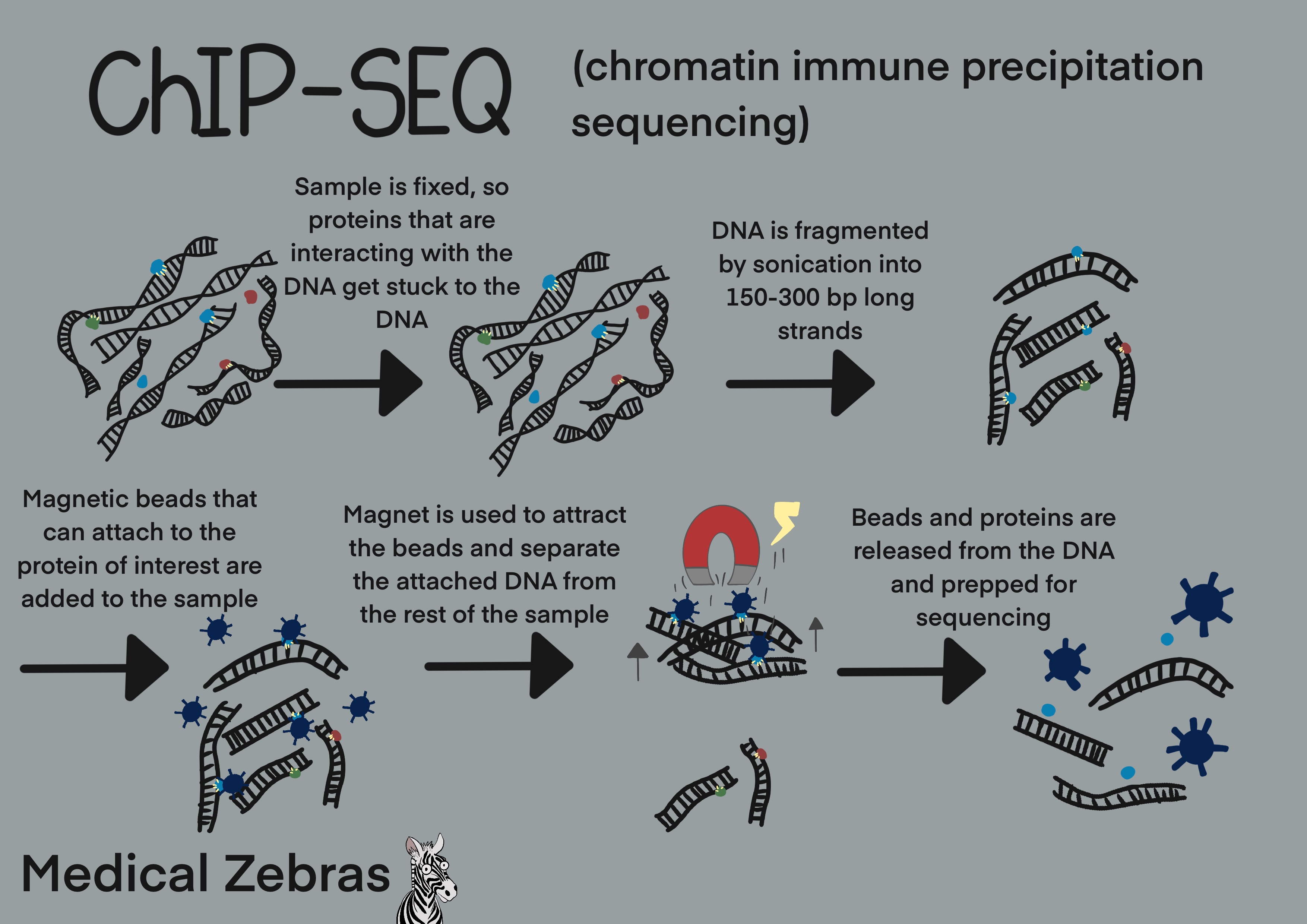
ChIP-Seq / CUT&RUN / CUT&Tag
ChIP(Chromatin Immunoprecipitation)-Seq is a method to identify binding
sites of DNA-associated proteins, most often transciption factors and
other proteins that regulate gene expression and therefore affect
phenotype.
First the sample is submerged in formaldehyde to cause crosslinking between the proteins that are in contact with DNA, and the DNA. Thereafter the DNA is fragmented by sonication and later cells are lysated (cell membrane destroyed) so we gain access to the crosslinked proteins and DNA. Next magnetic beads with an antibody that can attach to the protein of interest are added to the mix. The beads link to the protein of interest and by magnetic force we can isolate them from the rest of the sample. By reversing the crosslinking, we can now purify the DNA and later sequence it and thereby map where our gene of interest is located within the DNA.
As both formaldehyde and sonication can cause damage to the DNA, a good depth (sequencing reads) during sequencing is required to get reliable results. This increases the cost immensly and therefore newer techniques have been developed to try to avoid these costly factors.
One of the newer techniques to solve this issue is called CUT&RUN. In CUT &RUN the cells are immobilized on magnetic beads and permeabilized. Antibodies that target the protein of interest are then introduced to the sample and thereafter a Protein A-G Micrococcal nuclease is used to cut out the DNA that the antibody has attached to. Thereafter the released DNA is isolated and sequenced further. By using this micrococcal enzyme instead of fragmenting the DNA by sonication and avoiding using formaldehyde for crosslinking, background during sequencing is drastically reduced, meaning that the same depth during sequencing is not required, drastically decreasing sequencing cost.
An even newer technique has been developed similar to CUT&RUN, named CUT&Tag. Here instead of the micrococcal nuclease a transposon, specifically Protein A-Tn5 is used for cutting around the DNA that the antibody has attached to.
Simutaneously as the transposon cut it inserts adapters that are needed for further sequencing. By including adapter ligation in this step of the process preparations for sequencing are made simpler as one does only have to amplify the DNA before sequencing can start.
First the sample is submerged in formaldehyde to cause crosslinking between the proteins that are in contact with DNA, and the DNA. Thereafter the DNA is fragmented by sonication and later cells are lysated (cell membrane destroyed) so we gain access to the crosslinked proteins and DNA. Next magnetic beads with an antibody that can attach to the protein of interest are added to the mix. The beads link to the protein of interest and by magnetic force we can isolate them from the rest of the sample. By reversing the crosslinking, we can now purify the DNA and later sequence it and thereby map where our gene of interest is located within the DNA.
As both formaldehyde and sonication can cause damage to the DNA, a good depth (sequencing reads) during sequencing is required to get reliable results. This increases the cost immensly and therefore newer techniques have been developed to try to avoid these costly factors.
One of the newer techniques to solve this issue is called CUT&RUN. In CUT &RUN the cells are immobilized on magnetic beads and permeabilized. Antibodies that target the protein of interest are then introduced to the sample and thereafter a Protein A-G Micrococcal nuclease is used to cut out the DNA that the antibody has attached to. Thereafter the released DNA is isolated and sequenced further. By using this micrococcal enzyme instead of fragmenting the DNA by sonication and avoiding using formaldehyde for crosslinking, background during sequencing is drastically reduced, meaning that the same depth during sequencing is not required, drastically decreasing sequencing cost.
An even newer technique has been developed similar to CUT&RUN, named CUT&Tag. Here instead of the micrococcal nuclease a transposon, specifically Protein A-Tn5 is used for cutting around the DNA that the antibody has attached to.
Simutaneously as the transposon cut it inserts adapters that are needed for further sequencing. By including adapter ligation in this step of the process preparations for sequencing are made simpler as one does only have to amplify the DNA before sequencing can start.
Pros: With the newer techniques (CUT&RUN and CUT
&Tag), less cells are needed for good sequencing results (Chip-seq
~30million or more cells needed vs CUT&RUN where only over 5000 cells
are needed). Processing time is minimal and sequencing cost is reduced
as well.
Cons:
Examples of uses in the field
~ Not used in the clinical environment at the moment.
Cons:
Examples of uses in the field
~ Not used in the clinical environment at the moment.